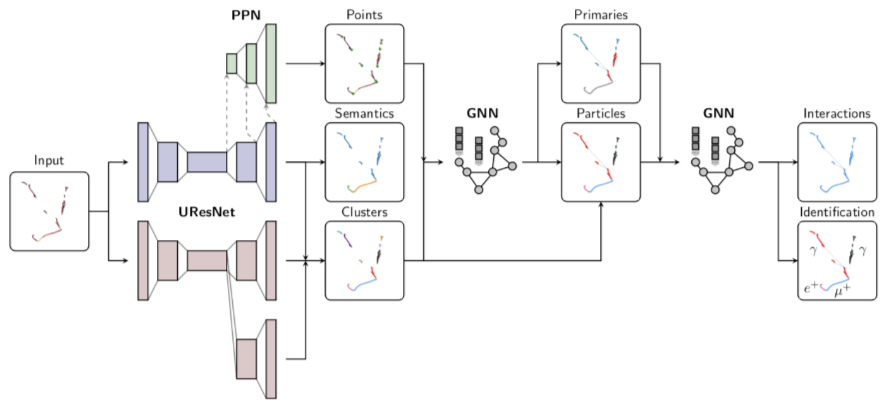
Particle clustering¶
Imports and configuration¶
If needed, you can edit the path to lartpc_mlreco3d library and to the data folder.
import os
SOFTWARE_DIR = '%s/lartpc_mlreco3d' % os.environ.get('HOME')
DATA_DIR = os.environ.get('DATA_DIR')
The usual imports and setting the right PYTHON_PATH… click if you need to see them.
import sys, os
# set software directory
sys.path.insert(0, SOFTWARE_DIR)
import numpy as np
import yaml
import torch
import plotly
import plotly.graph_objs as go
from plotly.offline import iplot, init_notebook_mode
init_notebook_mode(connected=False)
from mlreco.visualization import scatter_points, plotly_layout3d
from mlreco.visualization.gnn import scatter_clusters, network_topology, network_schematic
from mlreco.utils.ppn import uresnet_ppn_type_point_selector
from mlreco.utils.cluster.dense_cluster import fit_predict_np, gaussian_kernel
from mlreco.main_funcs import process_config, prepare
from mlreco.utils.gnn.cluster import get_cluster_label
from mlreco.utils.deghosting import adapt_labels_numpy as adapt_labels
from mlreco.visualization.gnn import network_topology
from mlreco.utils.cluster.cluster_graph_constructor import ClusterGraphConstructor
from larcv import larcv
/usr/local/lib/python3.8/dist-packages/MinkowskiEngine/__init__.py:36: UserWarning:
The environment variable `OMP_NUM_THREADS` not set. MinkowskiEngine will automatically set `OMP_NUM_THREADS=16`. If you want to set `OMP_NUM_THREADS` manually, please export it on the command line before running a python script. e.g. `export OMP_NUM_THREADS=12; python your_program.py`. It is recommended to set it below 24.
Welcome to JupyROOT 6.22/09
The configuration is loaded from the file inference.cfg.
cfg=yaml.load(open('%s/inference.cfg' % DATA_DIR, 'r').read().replace('DATA_DIR', DATA_DIR),Loader=yaml.Loader)
# pre-process configuration (checks + certain non-specified default settings)
process_config(cfg)
# prepare function configures necessary "handlers"
hs=prepare(cfg)
Config processed at: Linux ampt017 3.10.0-1160.42.2.el7.x86_64 #1 SMP Tue Sep 7 14:49:57 UTC 2021 x86_64 x86_64 x86_64 GNU/Linux
$CUDA_VISIBLE_DEVICES="0"
{ 'iotool': { 'batch_size': 10,
'collate_fn': 'CollateSparse',
'dataset': { 'data_keys': [ '/sdf/home/l/ldomine/lartpc_mlreco3d_tutorials/book/data/mpvmpr_062022_test_small.root'],
'limit_num_files': 10,
'name': 'LArCVDataset',
'schema': { 'cluster_label': [ 'parse_cluster3d_clean_full',
'cluster3d_pcluster',
'particle_pcluster',
'particle_mpv',
'sparse3d_pcluster_semantics'],
'input_data': [ 'parse_sparse3d_scn',
'sparse3d_reco',
'sparse3d_reco_chi2',
'sparse3d_reco_hit_charge0',
'sparse3d_reco_hit_charge1',
'sparse3d_reco_hit_charge2',
'sparse3d_reco_hit_key0',
'sparse3d_reco_hit_key1',
'sparse3d_reco_hit_key2'],
'kinematics_label': [ 'parse_cluster3d_kinematics_clean',
'cluster3d_pcluster',
'particle_corrected',
'particle_mpv',
'sparse3d_pcluster_semantics'],
'particle_graph': [ 'parse_particle_graph_corrected',
'particle_corrected',
'cluster3d_pcluster'],
'particles_asis': [ 'parse_particle_asis',
'particle_pcluster',
'cluster3d_pcluster'],
'particles_label': [ 'parse_particle_points_with_tagging',
'sparse3d_pcluster',
'particle_corrected'],
'segment_label': [ 'parse_sparse3d_scn',
'sparse3d_pcluster_semantics_ghost']}},
'minibatch_size': 10,
'num_workers': 1,
'shuffle': False},
'model': { 'loss_input': [ 'segment_label',
'particles_label',
'cluster_label',
'kinematics_label',
'particle_graph'],
'modules': { 'chain': { 'enable_charge_rescaling': True,
'enable_cnn_clust': True,
'enable_cosmic': False,
'enable_dbscan': True,
'enable_ghost': True,
'enable_gnn_inter': True,
'enable_gnn_kinematics': False,
'enable_gnn_shower': True,
'enable_gnn_track': True,
'enable_ppn': True,
'enable_uresnet': True,
'process_fragments': True,
'use_ppn_in_gnn': True,
'use_supp_in_gnn': True,
'use_true_fragments': False,
'verbose': True},
'cosmic_discriminator': { 'res_encoder': { 'coordConv': True,
'latent_size': 2,
'pool_mode': 'avg',
'spatial_size': 6144},
'use_input_data': False,
'use_true_interactions': False},
'cosmic_loss': { 'node_loss': { 'balance_classes': True,
'name': 'type',
'target_col': 8}},
'dbscan': { 'dbscan_fragment_manager': { 'cluster_classes': [ 0,
2,
3],
'delta_label': 3,
'eps': [ 1.999,
1.999,
4.999],
'michel_label': 2,
'num_classes': 4,
'ppn_score_threshold': 0.5,
'ppn_type_score_threshold': 0.5,
'track_clustering_method': 'masked_dbscan',
'track_label': 1}},
'graph_spice': { 'constructor_cfg': { 'cluster_col': 5,
'edge_cut_threshold': 0.1,
'edge_mode': 'attributes',
'hyper_dimension': 22,
'mode': 'knn',
'seg_col': -1},
'embedder_cfg': { 'graph_spice_embedder': { 'covariance_mode': 'softplus',
'feature_embedding_dim': 16,
'num_classes': 5,
'occupancy_mode': 'softplus',
'segmentationLayer': False,
'spatial_embedding_dim': 3},
'uresnet': { 'activation': { 'args': { 'negative_slope': 0.33},
'name': 'lrelu'},
'allow_bias': False,
'depth': 5,
'filters': 32,
'input_kernel': 5,
'norm_layer': { 'args': { 'eps': 0.0001,
'momentum': 0.01},
'name': 'batch_norm'},
'num_input': 4,
'reps': 2,
'spatial_size': 6144}},
'freeze_weights': True,
'kernel_cfg': { 'name': 'bilinear',
'num_features': 32},
'min_points': 3,
'node_dim': 22,
'skip_classes': [0, 2, 3, 4],
'use_raw_features': True,
'use_true_labels': False},
'graph_spice_loss': { 'edge_loss_cfg': { 'loss_type': 'LogDice'},
'eval': True,
'invert': True,
'kernel_lossfn': 'lovasz_hinge',
'name': 'graph_spice_edge_loss'},
'grappa_inter': { 'base': { 'add_start_dir': True,
'add_start_point': True,
'kinematics_mlp': True,
'kinematics_type': True,
'node_min_size': 3,
'node_type': [ 0,
1,
2,
3],
'start_dir_max_dist': 5,
'vertex_mlp': True},
'edge_encoder': { 'name': 'geo',
'use_numpy': True},
'gnn_model': { 'edge_classes': 2,
'edge_feats': 19,
'edge_output_feats': 64,
'name': 'meta',
'node_classes': 2,
'node_feats': 28,
'node_output_feats': 64},
'node_encoder': { 'name': 'geo',
'use_numpy': True},
'type_net': { 'num_hidden': 32},
'use_shower_primary': True,
'use_true_particles': False,
'vertex_net': { 'num_hidden': 32}},
'grappa_inter_loss': { 'edge_loss': { 'name': 'channel',
'source_col': 6,
'target_col': 7},
'node_loss': { 'balance_classes': True,
'name': 'kinematics',
'spatial_size': 6144}},
'grappa_kinematics': { 'base': { 'edge_dist_metric': 'set',
'edge_dist_numpy': True,
'edge_max_dist': -1,
'kinematics_mlp': True,
'kinematics_momentum': True,
'network': 'complete',
'node_min_size': -1,
'node_type': -1},
'edge_encoder': { 'cnn_encoder': { 'name': 'cnn',
'res_encoder': { 'coordConv': True,
'latent_size': 32,
'pool_mode': 'avg',
'spatial_size': 6144}},
'geo_encoder': { 'more_feats': True},
'name': 'mix_debug',
'normalize': True},
'gnn_model': { 'edge_classes': 2,
'edge_feats': 51,
'edge_output_feats': 64,
'leak': 0.33,
'name': 'nnconv_old',
'node_classes': 5,
'node_feats': 83,
'node_output_feats': 128},
'momentum_net': { 'num_hidden': 32},
'node_encoder': { 'cnn_encoder': { 'name': 'cnn',
'res_encoder': { 'coordConv': True,
'input_kernel': 3,
'latent_size': 64,
'pool_mode': 'avg',
'spatial_size': 6144}},
'geo_encoder': { 'more_feats': True},
'name': 'mix_debug',
'normalize': True},
'use_true_particles': False},
'grappa_kinematics_loss': { 'edge_loss': { 'name': 'channel',
'target': 'particle_forest'},
'node_loss': { 'name': 'kinematics',
'reg_loss': 'l2'}},
'grappa_shower': { 'base': { 'add_start_dir': True,
'add_start_point': True,
'node_min_size': -1,
'node_type': 0,
'start_dir_max_dist': 5},
'edge_encoder': { 'name': 'geo',
'use_numpy': True},
'freeze_weights': True,
'gnn_model': { 'edge_classes': 2,
'edge_feats': 19,
'edge_output_feats': 64,
'name': 'meta',
'node_classes': 2,
'node_feats': 28,
'node_output_feats': 64},
'node_encoder': { 'name': 'geo',
'use_numpy': True}},
'grappa_shower_loss': { 'edge_loss': { 'high_purity': True,
'name': 'channel',
'source_col': 5,
'target_col': 6},
'node_loss': { 'high_purity': True,
'name': 'primary',
'use_group_pred': True}},
'grappa_track': { 'base': { 'add_start_dir': True,
'add_start_point': True,
'node_min_size': 3,
'node_type': 1,
'start_dir_max_dist': 5},
'edge_encoder': { 'name': 'geo',
'use_numpy': True},
'freeze_weights': True,
'gnn_model': { 'edge_classes': 2,
'edge_feats': 19,
'edge_output_feats': 64,
'name': 'meta',
'node_classes': 2,
'node_feats': 28,
'node_output_feats': 64},
'node_encoder': { 'name': 'geo',
'use_numpy': True}},
'grappa_track_loss': { 'edge_loss': { 'name': 'channel',
'source_col': 5,
'target_col': 6}},
'uresnet_deghost': { 'freeze_weights': True,
'uresnet_lonely': { 'activation': { 'args': { 'negative_slope': 0.33},
'name': 'lrelu'},
'allow_bias': False,
'depth': 5,
'filters': 32,
'ghost': False,
'norm_layer': { 'args': { 'eps': 0.0001,
'momentum': 0.01},
'name': 'batch_norm'},
'num_classes': 2,
'num_input': 2,
'reps': 2,
'spatial_size': 6144}},
'uresnet_ppn': { 'ppn': { 'classify_endpoints': True,
'depth': 5,
'filters': 32,
'freeze_weights': True,
'mask_loss_name': 'BCE',
'num_classes': 5,
'particles_label_seg_col': -3,
'ppn_resolution': 1.0,
'ppn_score_threshold': 0.6,
'spatial_size': 6144},
'uresnet_lonely': { 'activation': { 'args': { 'negative_slope': 0.33},
'name': 'lrelu'},
'allow_bias': False,
'depth': 5,
'filters': 32,
'freeze_weights': True,
'norm_layer': { 'args': { 'eps': 0.0001,
'momentum': 0.01},
'name': 'batch_norm'},
'num_classes': 5,
'num_input': 2,
'reps': 2,
'spatial_size': 6144}}},
'name': 'full_chain',
'network_input': [ 'input_data',
'segment_label',
'cluster_label']},
'trainval': { 'checkpoint_step': 100,
'concat_result': [ 'input_edge_features',
'input_node_features',
'points',
'coordinates',
'particle_node_features',
'particle_edge_features',
'track_node_features',
'shower_node_features',
'ppn_coords',
'mask_ppn',
'ppn_layers',
'classify_endpoints',
'vertex_layers',
'vertex_coords',
'primary_label_scales',
'segment_label_scales',
'seediness',
'margins',
'embeddings',
'fragments',
'fragments_seg',
'shower_fragments',
'shower_edge_index',
'shower_edge_pred',
'shower_node_pred',
'shower_group_pred',
'track_fragments',
'track_edge_index',
'track_node_pred',
'track_edge_pred',
'track_group_pred',
'particle_fragments',
'particle_edge_index',
'particle_node_pred',
'particle_edge_pred',
'particle_group_pred',
'particles',
'inter_edge_index',
'inter_node_pred',
'inter_edge_pred',
'inter_group_pred',
'inter_particles',
'node_pred_p',
'node_pred_type',
'kinematics_node_pred_p',
'kinematics_node_pred_type',
'flow_edge_pred',
'kinematics_particles',
'kinematics_edge_index',
'clust_fragments',
'clust_frag_seg',
'interactions',
'inter_cosmic_pred',
'node_pred_vtx',
'total_num_points',
'total_nonghost_points',
'spatial_embeddings',
'occupancy',
'hypergraph_features',
'features',
'feature_embeddings',
'covariance',
'clusts',
'edge_index',
'edge_pred',
'node_pred'],
'debug': False,
'gpus': [0],
'iterations': 10,
'log_dir': './log_trash',
'minibatch_size': -1,
'model_path': '/sdf/home/l/ldomine/lartpc_mlreco3d_tutorials/book/data/weights_full_mpvmpr_062022.ckpt',
'optimizer': {'args': {'lr': 0.001}, 'name': 'Adam'},
'report_step': 1,
'seed': 123,
'train': False,
'unwrapper': 'unwrap_3d_mink',
'weight_prefix': './weights_trash/snapshot'}}
Loading file: /sdf/home/l/ldomine/lartpc_mlreco3d_tutorials/book/data/mpvmpr_062022_test_small.root
Loading tree sparse3d_reco
Loading tree sparse3d_reco_chi2
Loading tree sparse3d_reco_hit_charge0
Loading tree sparse3d_reco_hit_charge1
Loading tree sparse3d_reco_hit_charge2
Loading tree sparse3d_reco_hit_key0
Loading tree sparse3d_reco_hit_key1
Loading tree sparse3d_reco_hit_key2
Loading tree sparse3d_pcluster_semantics_ghost
Loading tree cluster3d_pcluster
Loading tree particle_pcluster
Loading tree particle_mpv
Loading tree sparse3d_pcluster_semantics
Loading tree sparse3d_pcluster
Loading tree particle_corrected
Found 101 events in file(s)
Shower GNN: True
Track GNN: True
Particle GNN: False
Interaction GNN: True
Kinematics GNN: False
Cosmic GNN: False
Since one of the GNNs are turned on, process_fragments is turned ON.
Fragment processing is turned ON. When training CNN models from
scratch, we recommend turning fragment processing OFF as without
reliable segmentation and/or cnn clustering outputs this could take
prohibitively large training iterations.
Shower GNN: True
Track GNN: True
Particle GNN: False
Interaction GNN: True
Kinematics GNN: False
Cosmic GNN: False
Since one of the GNNs are turned on, process_fragments is turned ON.
Fragment processing is turned ON. When training CNN models from
scratch, we recommend turning fragment processing OFF as without
reliable segmentation and/or cnn clustering outputs this could take
prohibitively large training iterations.
Freezing 82 weights for a sub-module ppn
Freezing 141 weights for a sub-module uresnet_lonely
Freezing 141 weights for a sub-module uresnet_deghost
Freezing 146 weights for a sub-module graph_spice
Freezing 120 weights for a sub-module grappa_track
Freezing 120 weights for a sub-module grappa_shower
Restoring weights for from /sdf/home/l/ldomine/lartpc_mlreco3d_tutorials/book/data/weights_full_mpvmpr_062022.ckpt...
Done.
Warning in <TClass::Init>: no dictionary for class larcv::EventNeutrino is available
Warning in <TClass::Init>: no dictionary for class larcv::NeutrinoSet is available
Warning in <TClass::Init>: no dictionary for class larcv::Neutrino is available
The output is hidden because it reprints the entire (lengthy) configuration. Feel free to take a look if you are curious!
Finally we run the chain for 1 iteration:
# Call forward to run the net, store the output in "res"
data, output = hs.trainer.forward(hs.data_io_iter)
Deghosting Accuracy: 0.9830
Segmentation Accuracy: 0.9900
PPN Accuracy: 0.8843
Clustering Accuracy: 0.2691
Clustering Edge Accuracy: 0.1252
Shower fragment clustering accuracy: 0.9581
Shower primary prediction accuracy: 0.9434
Track fragment clustering accuracy: 0.9937
Interaction grouping accuracy: 0.9763
Particle ID accuracy: 0.8409
Primary particle score accuracy: 0.9755
Now we can play with data and output to visualize what we are interested in. Feel free to change the
entry index if you want to look at a different entry!
entry = 0
Let us grab the interesting quantities:
clust_label = data['cluster_label'][entry]
input_data = data['input_data'][entry]
segment_label = data['segment_label'][entry][:, -1]
ghost_mask = output['ghost'][entry].argmax(axis=1) == 0
segment_pred = output['segmentation'][entry].argmax(axis=1)
Track clustering¶
Stage 1: GraphSPICE¶
GraphSPICE is a first pass of CNN-based voxel clustering. We optimize this round for purity, hence there might be many small track fragments predicted by this pass.
graph = output['graph'][0]
graph_info = output['graph_info'][0]
gs_manager = ClusterGraphConstructor(cfg['model']['modules']['graph_spice']['constructor_cfg'], graph_batch=graph, graph_info=graph_info)
track_label = 1
Time to run the fit function that looks at the predicted embeddings and does the actual clustering inference for us:
pred, G, subgraph = gs_manager.fit_predict_one(entry)
And visualization time!
trace = []
edep = input_data[segment_label < 5]
trace+= scatter_points(clust_label[clust_label[:, -1] == track_label],markersize=1,color=clust_label[clust_label[:, -1] == track_label, 6], cmin=0, cmax=50, colorscale=plotly.colors.qualitative.D3)
trace[-1].name = 'True cluster labels (true no-ghost mask)'
clust_label_adapted = adapt_labels(output, data['segment_label'], data['cluster_label'])[entry]
trace+= scatter_points(clust_label_adapted[segment_pred[ghost_mask] == track_label],markersize=1,color=clust_label_adapted[segment_pred[ghost_mask] == track_label, 6], cmin=0, cmax=50, colorscale=plotly.colors.qualitative.D3)
trace[-1].name = 'True cluster labels (predicted no-ghost mask & semantic)'
trace+= scatter_points(subgraph.pos.cpu().numpy(),markersize=1,color=pred, cmin=0, cmax=50, colorscale=plotly.colors.qualitative.D3)
trace[-1].name = 'Predicted clusters (predicted no-ghost mask)'
# trace+= scatter_points(clust_label,markersize=1,color=output['graph_spice_label'][entry][:, 5], cmin=0, cmax=10, colorscale=plotly.colors.qualitative.D3)
# trace[-1].name = 'Input to graph spice'
fig = go.Figure(data=trace,layout=plotly_layout3d())
fig.update_layout(legend=dict(x=1.0, y=0.8))
iplot(fig)
Stage 2: GNN¶
We use a GNN to then cluster these small track fragments together. First we need to retrieve true labels that take into account the ghost points. Labels are assigned by nearest non-ghost voxel.
clust_label_adapted = adapt_labels(output, data['segment_label'], data['cluster_label'])[entry]
clust_ids_true = get_cluster_label(torch.tensor(clust_label_adapted), output['track_fragments'][entry], column=6)
clust_ids_pred = output['track_group_pred'][entry]
Then we can look at the visualization:
trace = []
trace += network_topology(data['input_data'][entry][ghost_mask],
output['track_fragments'][entry],
#edge_index=output['frag_edge_index'][entry],
clust_labels=clust_ids_true,
markersize=2, cmin=0, cmax=40, colorscale=plotly.colors.qualitative.Dark24)
trace[-1].name = 'Cluster truth'
trace += network_topology(data['input_data'][entry][ghost_mask],
output['track_fragments'][entry],
#edge_index=output['frag_edge_index'][entry],
clust_labels=clust_ids_pred,
markersize=2, cmin=0, cmax=40, colorscale=plotly.colors.qualitative.Dark24)
trace[-1].name = 'Cluster predictions'
fig = go.Figure(data=trace,layout=plotly_layout3d())
#fig = go.Figure(data=trace,layout=layout)
fig.update_layout(legend=dict(x=1.1, y=0.9))
iplot(fig)
Note that this uses a different helper function from lartpc_mlreco3d: the function
network_topology is useful to visualize the output of a GNN. Its first argument is
the input data (to provide the voxel coordinates), its second argument is a list of
list of voxel indices (list of fragments).
Shower clustering¶
clust_label_adapted = adapt_labels(output, data['segment_label'], data['cluster_label'])[entry]
clust_ids_true = get_cluster_label(torch.tensor(clust_label_adapted), output['shower_fragments'][entry], column=6)
clust_ids_pred = output['shower_group_pred'][entry]
trace = []
trace += network_topology(data['input_data'][entry][ghost_mask],
output['shower_fragments'][entry],
#edge_index=output['frag_edge_index'][entry],
clust_labels=clust_ids_true,
markersize=2, cmin=0, cmax=10, colorscale=plotly.colors.qualitative.Dark24)
trace[-1].name = 'Cluster truth'
trace += network_topology(data['input_data'][entry][ghost_mask],
output['shower_fragments'][entry],
#edge_index=output['frag_edge_index'][entry],
clust_labels=clust_ids_pred,
markersize=2, cmin=0, cmax=10, colorscale=plotly.colors.qualitative.Dark24)
trace[-1].name = 'Cluster predictions'
fig = go.Figure(data=trace,layout=plotly_layout3d())
fig.update_layout(legend=dict(x=1.1, y=0.9))
iplot(fig)